Insights on molecular level
iMScope TRIO with revolutionary technology
In recent years, imaging mass spectrometry (imaging MS) has received increased attention through a variety of applications. This fairly new innovation in mass spectrometry allows spatial assignment of analytes as well as their identification. The sample – for instance, a tissue section – is analyzed using mass spectrometry at specified locations in a grid. The mass spectra of the individual points are subsequently combined via dedicated software into one image (MS image) which represents the distribution of individual substances within the sample. This way, the occurrence of certain molecules at specific locations can be compared with morphological abnormalities.
With the iMScope TRIO (Figure 1), Shimadzu offers a worldwide unique instrument for imaging mass spectrometry. The iMScope combines an optical microscope and a high-resolution mass spectrometer in one single instrument. This allows direct comparison of the MS image with the microscopic image. Substances from samples that were microscopically examined cannot only be identified directly, but their spatial distribution within a tissue can also be determined.
 iMScope TRIO – Imaging Mass Microscope
iMScope TRIO – Imaging Mass Microscope
World’s best spatial resolution
The optical microscope can acquire images in epi- and trans-illumination, as well as fluorescence images (up to 5 optional wavelengths). The analytes are ionized via MALDI (Matrix-Assisted Laser Desorption/Ionization) at atmospheric pressure. The solid-state laser used for this purpose can scan the sample with a variable diameter of 5 to 200 µm. This results in a maximum resolution of the MS images of 5 µm which is currently the world’s best spatial resolution that can be achieved.
The ionized molecules are subsequently analyzed using the integrated ion-trap time-of-flight (IT-TOF) mass spectrometer. The high-resolution system can also carry out MSn analyses which enables identification as well as structural elucidation of the target analytes. Furthermore, MS/MS analysis increases the sensitivity due to a better signal-to-noise ratio.
With an analysis speed of six pixels per second, MS images can be acquired rapidly. This way, acquisition of 250 x 250 pixels (62,500 pixels) takes less than three hours. The possibility of exchanging the MALDI source for an ESI (Electrospray Ionization) source further increases the flexibility of the system and the mass spectrometer can also be used for the analysis of liquid samples.
Large application bandwidth
There is a wide range of possible application areas, such as the analysis of biomarkers, active substances and their metabolites in tissues, the distribution of substances in foods and plants, but also the analysis of micro-defects or minute contaminations in synthetic materials. In general, the iMScope TRIO is used in innovative Research & Development.
The following example shows which additional information is gained from the comparison of an optical image with an MS image. A rat liver section was treated with quercetin and used for analysis. The MS image (Figure 1a) alone shows an apparently random uniform distribution of quercetin over the entire sample. After overlaying with the microscope image (Figure 1b), a correlation between the localization of the molecule and the structures within the investigated liver tissue can already be visualized. Magnification of the overlayed image (Figure 3) clearly shows that all the detected quercetin molecules are exclusively localized in the intracellular space or on the cell walls and not inside the cells.
 Figure 1a: MS/MS image of quercetin (m/z 269.2 -> 224.97)
Figure 1a: MS/MS image of quercetin (m/z 269.2 -> 224.97)
 Figure 1b: Overlay of the MS/MS image with the optical image
Figure 1b: Overlay of the MS/MS image with the optical image
The use of a larger laser beam diameter would have resulted in a loss of spatial information about the localization of quercetin within the tissue. A precise allocation to the intracellular space or the cell membrane would not have been possible.
 Figure 2: By enlarging the superimposed image, it can be seen that quercetin is localized in the intracellular space and not inside the cells
Figure 2: By enlarging the superimposed image, it can be seen that quercetin is localized in the intracellular space and not inside the cells
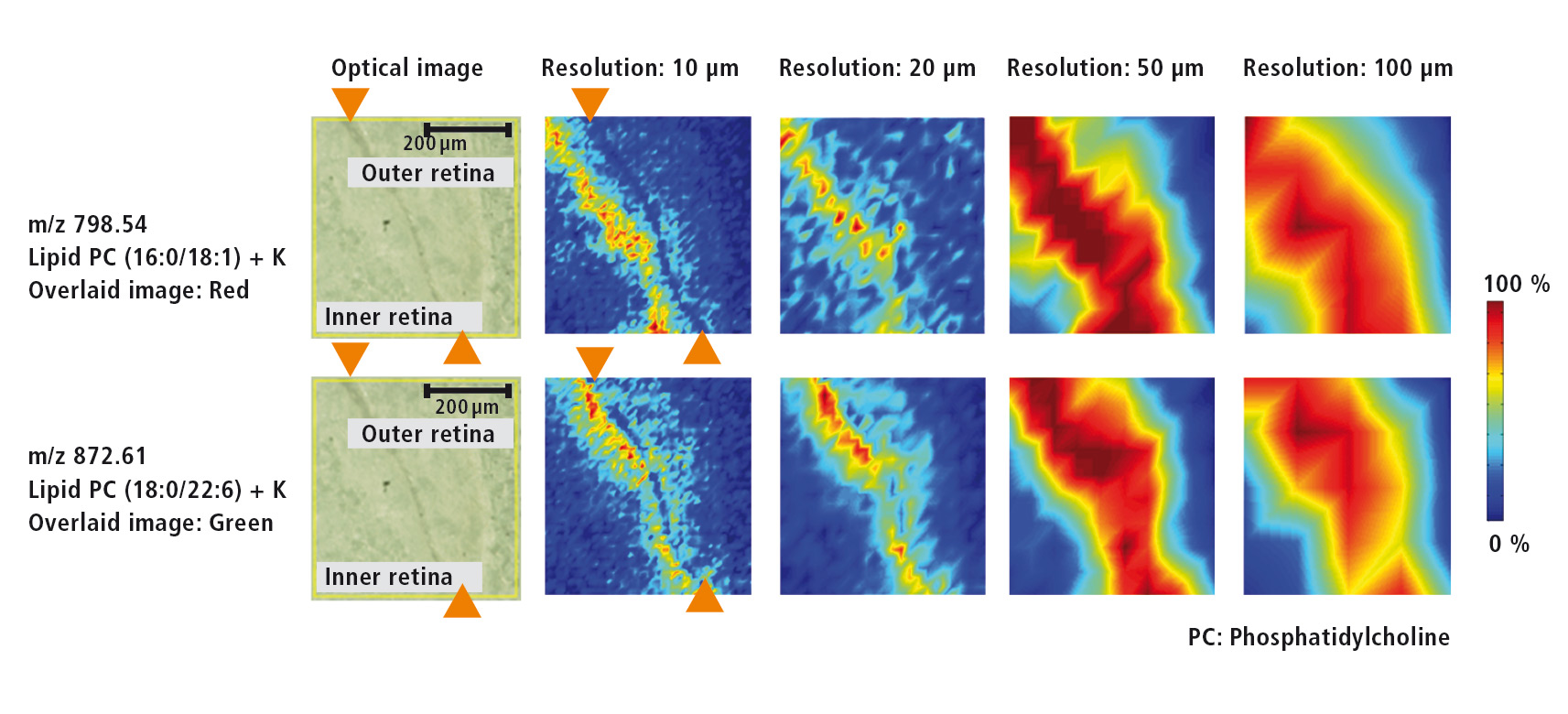 Figure 3: Microscopic image of the retina as well as MS images for two different lipids at resolutions of each 10 µm, 20 µm, 50 µm and 100 µm
Figure 3: Microscopic image of the retina as well as MS images for two different lipids at resolutions of each 10 µm, 20 µm, 50 µm and 100 µm
Resolution is application dependent
Depending on the investigated object, a medium or high spatial resolution is required. In order to study the localization within larger tissue structures, a medium resolution is sufficient, whereas localization on the molecular level requires the highest possible resolution.
High resolutions can be used to determine the distribution of drugs in microstructures or the accumulation of their metabolites.
For the investigation of various lipids that are present in the retina of the eye, a higher resolution of 10 µm was selected. The microscopic image (Figure 4) shows the retinal pigment epithelium as a dark band that separates the inner from the outer retina. MS images (Figure 4) of two different lipids (m/z = 798.54 und m/z = 872.61) were acquired using several resolutions (10 µm, 20 µm, 50 µm und 100 µm). The outline of the retinal pigment epithelium only can be recognized at a resolution of at least 10 µm. At a lower resolution this outline is lost.
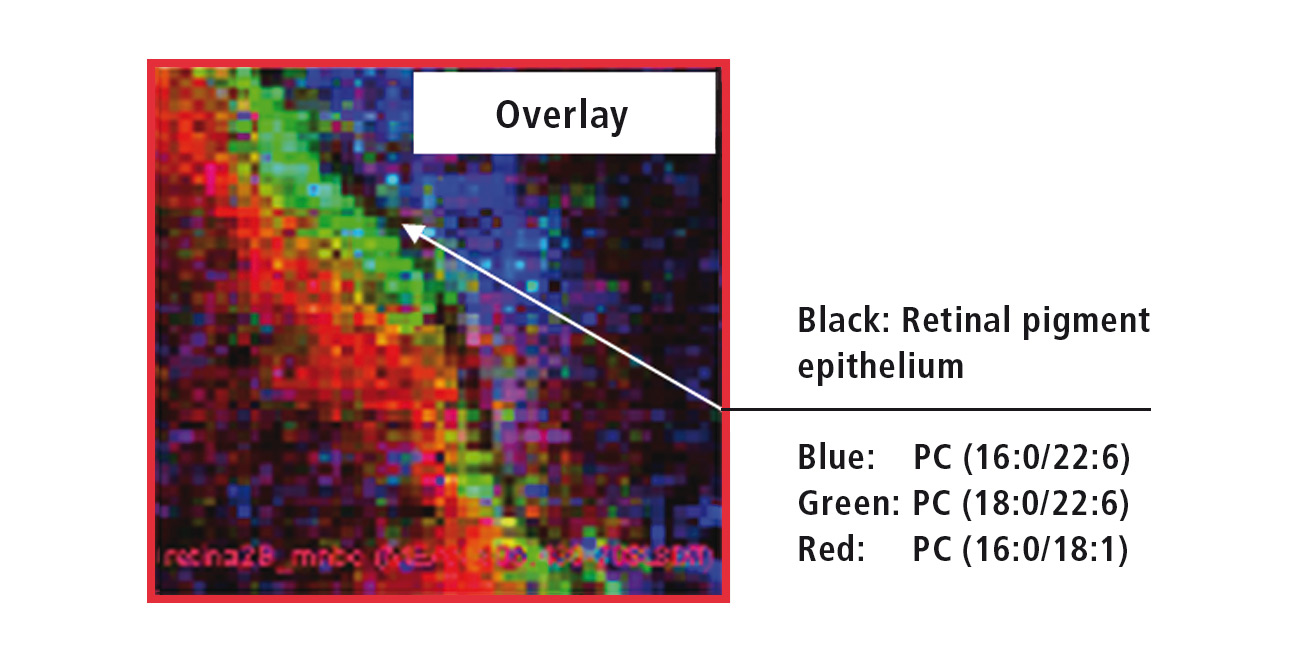 Figure 4: Overlay of the MS images from three different retinal lipids
Figure 4: Overlay of the MS images from three different retinal lipids
Software for data acquisition and analysis
The ‘Imaging MS solution’ software specifically developed for the iMScope TRIO is used for data acquisition as well as for data analysis. The software allows the overlaying of several MS images (Figure 5). The high resolution of the MS images allows for accurate assignment of the three investigated lipids. While PC (phosphatidylcholine, 16:0/22:6) exclusively occurs in the outer retina, PC (18:0/22:6) can be found directly adjacent to the retinal pigment epithelium in the inner retina, and PC (16:0/18:1) even further inside the retina.
Additional tools for statistical data analysis are Hierarchical Cluster Analysis (HCA), Region of Interest Analysis (ROI) and Principal Component Analysis (PCA).
Matrix deposition without delocalization
To obtain MS images with maximum spatial resolution, sample preparation is very important, next to an excellent performance of the mass spectrometer. The matrix required for ionization is conventionally dissolved in liquid and sprayed or spotted onto the sample. As a result, there is always a risk that the target analytes will dissolve in this liquid and diffuse inside the sample. This will lead to the situation that despite using instruments, which are capable of high-resolution sample analysis, the sample no longer yields such a high resolution.
With the iMLayer, Shimadzu therefore offers a device for matrix application with which this effect cannot occur. The matrix is deposited via a sublimation process, whereby a thin layer of very small matrix crystals is applied, and delocalization of analytes cannot occur. In combination with the iMLayer, the iMScope offers a unique and complete solution for sample preparation, acquisition and overlay of optical and MS images with the world’s best spatial resolution, as well as (statistical) data evaluation via imaging MS solution software.