Together we are strong

The demand to identify and characterize unknown substances occurs in many application fields in the pharmaceutical industry, for example analysis of impurities of synthesized compounds, degradation products in stability studies, metabolite identification to detect expected and unexpected metabolites and metabolite profiling experiment to search for differences in mass profiles in larger sample sets and their identification. In general, identification is an important part, because further actions are based on this identification. The ability to deliver an answer in a reasonable time or to efficiently support the decision process for further characterization steps defines the strength of a total solution.
LCMS-IT-TOF with its many unique software options is one of such powerful solutions.
Combining two different mass spectrometric tools (ion trap and TOF in one instrument) results in capabilities beyond any single mass analyzing technology (Figure 1). Ion trap mass spectrometry allows target molecules to be selected and fragmented easily (MSn; MS/MS, MS/MS/MS, MS/ MS/MS/MS…) while time-of-flight MS provides high mass resolution and accurate mass determination.
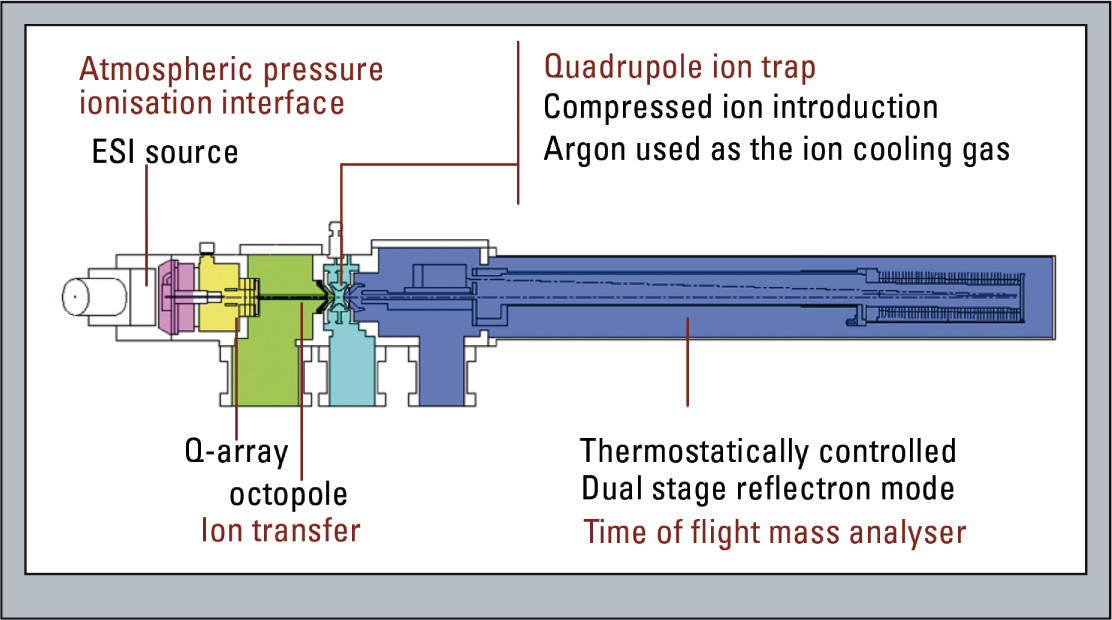 Figure 1: Schematic of the LCMS-IT-TOF
Figure 1: Schematic of the LCMS-IT-TOF
The LCMS-IT-TOF can be equipped with different ionization sources (ESI, APCI, APPI, ESI/ APCI combi source as well as a nano spray source) and can easily be combined with fast chromatographic separation delivering the required results with 100 msec polarity switching time in one run. In general, it delivers a mass accuracy of around 3 ppm externally calibrated, independent of the MS modus. A flight tube constantly kept at 40° Celsius supports mass stability even during analysis of larger sample set studies.
The easy and reliable generation of high quality data is an important issue for identifying unknown compounds. However, the software tools supporting interpretation of the data are equally of importance.
In addition to the fragment information, the elemental composition and the isotopic pattern of a compound are important information for the characterization of an unknown substance. Recent publications have highlighted that mass accuracy, while extremely important, is not sufficient for confident compound confirmation and identification of unknowns. Accurate isotopic abundances of measured ions are critical as well. The relative isotope abundance error has to be below 5 % for effective molecular formula generation for unknowns, helping to eliminate a large percentage of possible compounds.
“Formula Predictor” software
To determine the elemental composition of an unknown sample, Shimadzu’s Formula Predictor software uses all generated accurate masses of the compound, including the parent mass as well as generated fragment masses from the MSn experiments to calculate and validate the total elemental composition of the molecule of interest, and generates a candidate list. Additionally, the software uses isotope pattern information to compare the measured sample with the theoretical pattern of potential candidates which either increases the probability score or excludes certain candidates from the ranking. This dramatically decreases the number of false positive candidates and makes determination much easier for the operator. Shimadzu holds the patent for using the fragment information of MSn experiments to predict the elemental composition of the molecular ion.
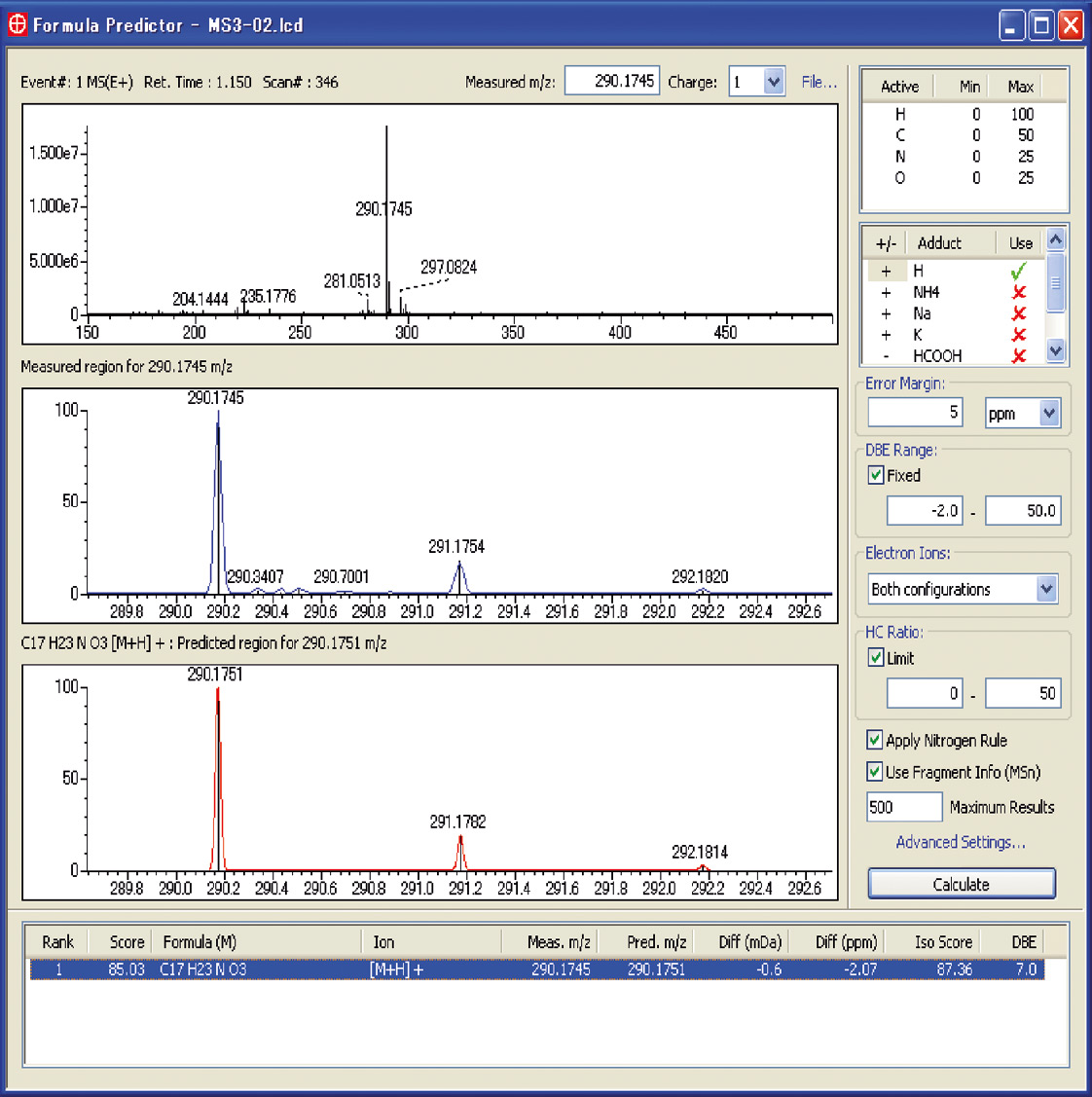 Figure 2: Formula Predictor software window showing one hit after using MS3 spectra
Figure 2: Formula Predictor software window showing one hit after using MS3 spectra
MetID Solution identifies metabolites in time-series analyses of bio fluids
The MetID Solution is used in the field of toxicity assessment/toxicology, where pharmaceutical companies test the toxicity of potential drug candidates; it is applied in the field of functional genomics where it determines the phenotype caused by a genetic manipulation, such as gene deletion or insertion. The MetID Solution software combines three different approaches to determine expected as well as unexpected metabolites.
First Workflow – Traditional Peak Picking
One or more time-series analyte files (i.e. subject’s biofluid at intervals after taking drug) are screened against a control file (i.e. subject’s biofluid just after taking drug). The common peaks are excluded, the remaining peaks contain metabolites.
There are published biotransformations for which parent drug functional groups can be metabolized (added to or replaced by) in the body. These peaks are called expected transformations and can be screened for by generating accurate mass XIC’s (Extracted Ion Chromatogram) using the metabolized formula.
Remaining peaks that cannot be categorized are called unexpected and are manually checked for metabolic likelihood. This can be laborious, but the Formula Predictor software reduces the effort.
Second workflow – Isotope Labelling
To aid confirmation of metabolites, discovery labs can label the parent using rare isotopes (which may or not be radioactive).
MetID automatically handles labelling in the Peak Picking workflow and can also use peaks in a radiolabel detector chromatogram.
Additionally, it can synthesize an Isotope Filtered Chromatogram from the MS chromatogram to locate these labelled metabolites (which helps to locate compounds containing distinctive isotope patterns such as halogens; schematic shown in figure 3).
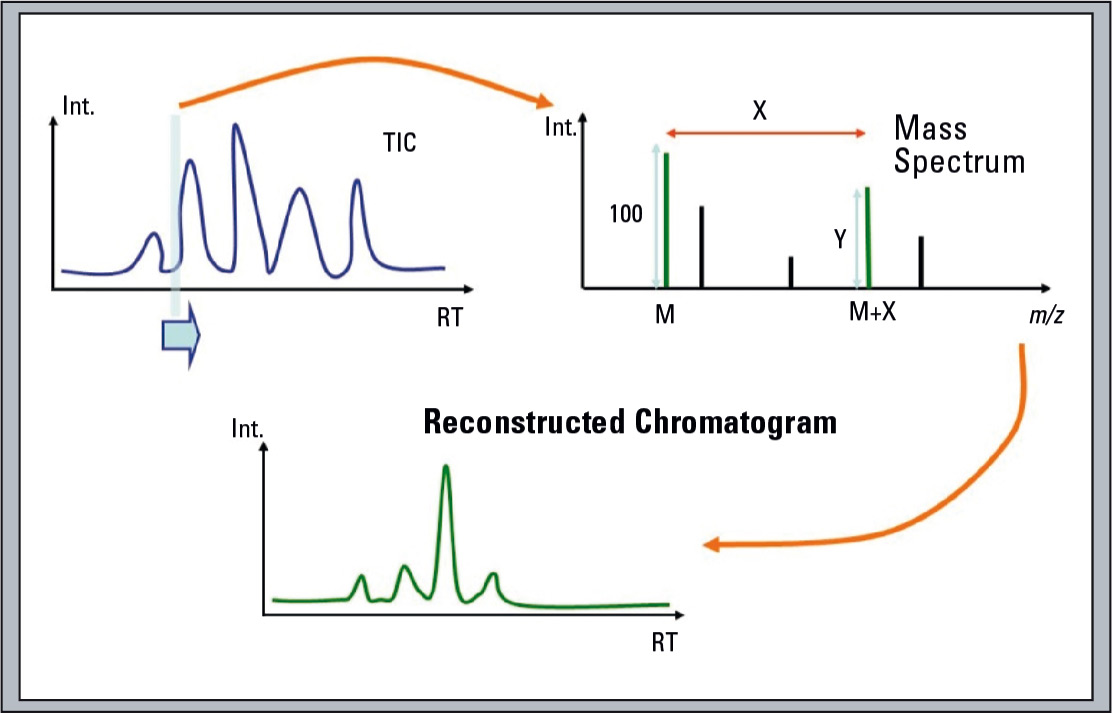 Figure 3: Schematic of the Isotope Filtering Chromatogram procedure
Figure 3: Schematic of the Isotope Filtering Chromatogram procedure
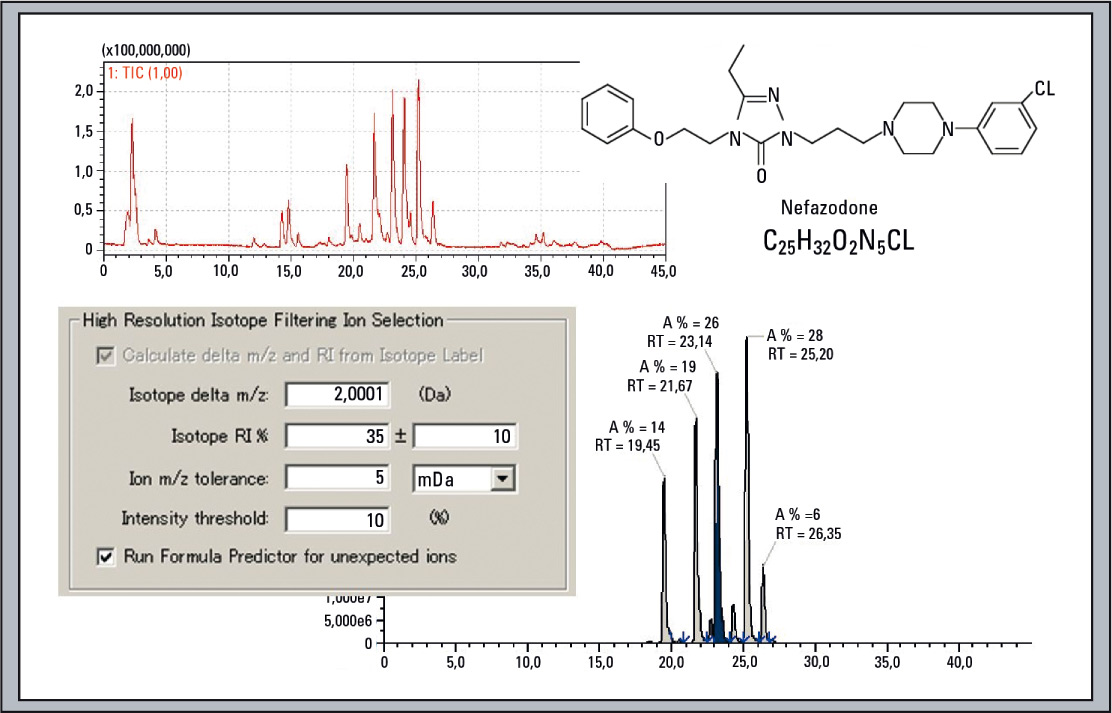 Figure 4: Real example of extracted isotope filtered chromatogram of Nefazodone, based on the chlorine isotope distribution
Figure 4: Real example of extracted isotope filtered chromatogram of Nefazodone, based on the chlorine isotope distribution
Third workflow – PLS regression analysis (Partial least square)
With the LCMS-IT-TOF the constant mass accuracy and resolution of MSn data can be utilized to first build a ‘fingerprint’ of the parent compound or drug (Y) and then acquire MSn data for each peak during analysis to get metabolite ‘fingerprints’ (X’s).
Partial least squares regression scores show how similar each X is to Y and since metabolites share functional groups with the parent, many unexpected peaks can be screened rapidly for potential metabolites and also to find new biotransformations.
Profiling Solution software
The comparison of large data sets and the detection of difference is the strength of the Profiling Solution software package. Fast and reliable, it uses the generated chromatographic and MS data of multiple sample measurements to recognize a difference (Figure 5).
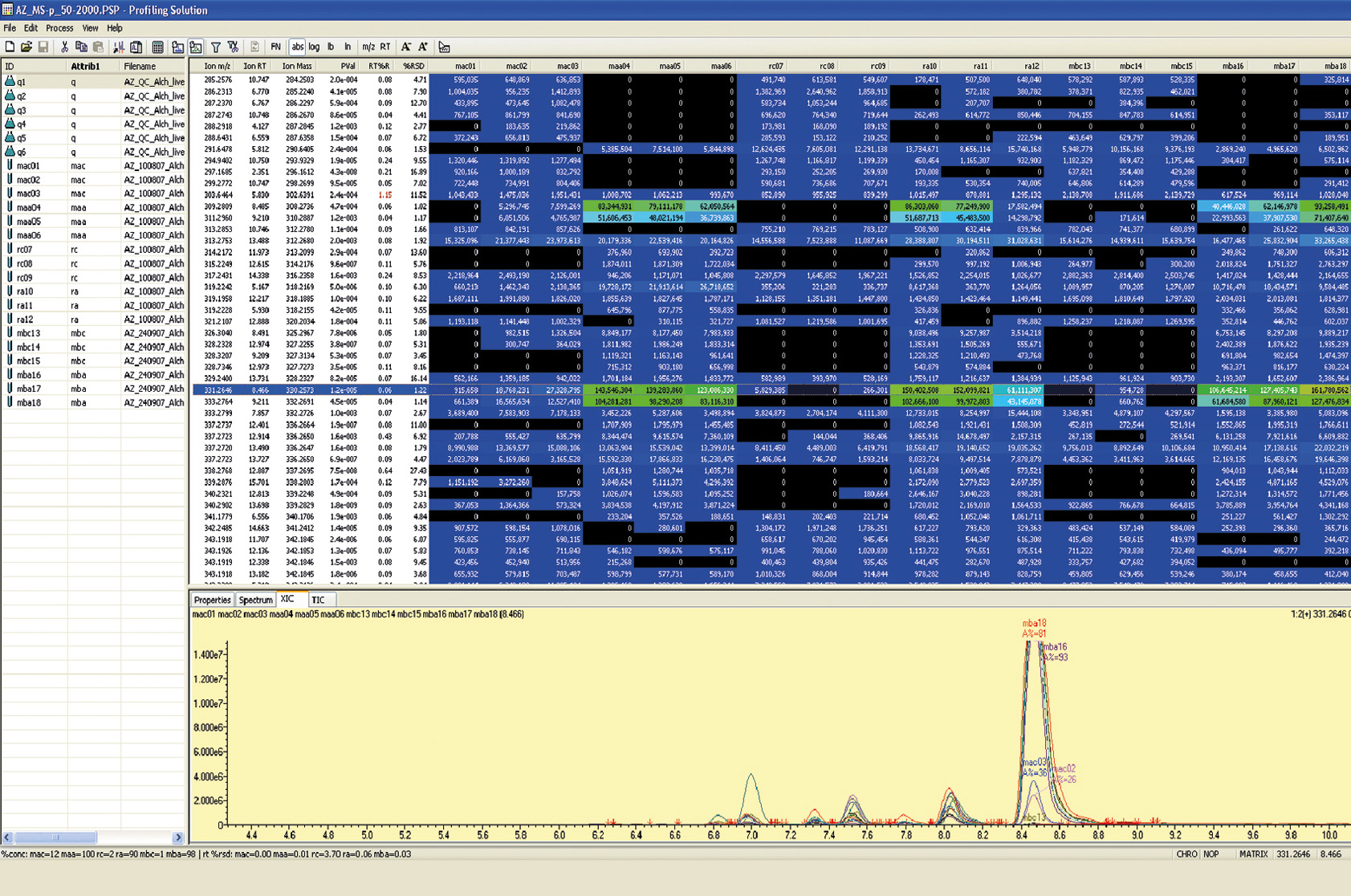 Figure 5: Array of different samples where significant variations are highlighted
Figure 5: Array of different samples where significant variations are highlighted
The software supports and offers the following steps:
- In a patented way, it aligns multiple data sets using a unique spectral binning technique and generates an aligned data array using spectrum data (mass resolution independent) and chromatogram data for multiple data streams (supports polarity switching within a run)
- Integrated filters for pooled QC analysis (filtering out idiosyncratic ion behaviour based on peak area, retention time)
- Integrated statistical tools (p-value, normalisation, de-isotoping)
- Export to statistical packages (SIMCA-P, MatLab as a text file)
- Hot links to web based data search engines (LipidMaps, KEGG, Metlin, ChemSpider)
Open Solution ComponentID
In many synthesis laboratories chemists get access to a single quadrupole system using “Open Access” software. However, in order to get an accurate mass measurement for a target compound the mass spectrometry department in general have to analyze the samples. This can create a big overhead for this mass spectrometry service departments. Open Solution Component ID is a software tool to automate the formula prediction workflow (Figure 6a). Its combination with the reliability of the LCMS-IT-TOF makes it to a powerful solution for molecular weight confirmation and impurities profiling.
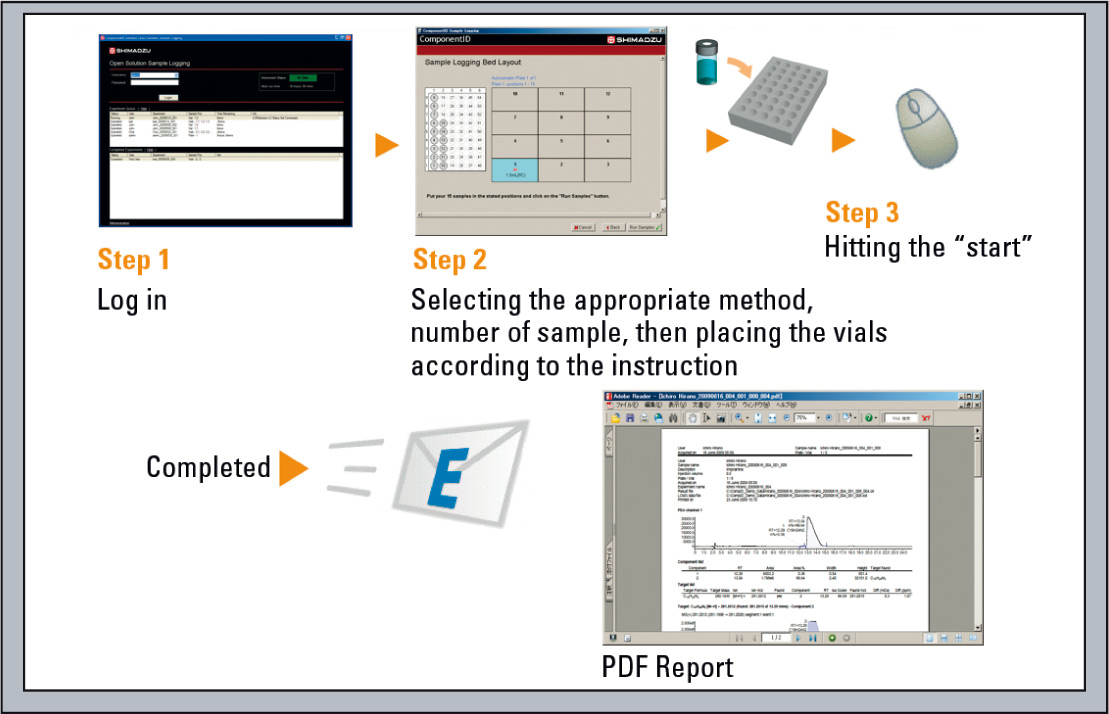 Figure 6a: Workflow to generate accurate mass data in an open access environment
Figure 6a: Workflow to generate accurate mass data in an open access environment
ComponentID provides a simple and easy to understand user interface for logging samples (and running analyses) and printing reports of results using the LCMS-IT-TOF (Figure 6b). This makes it easy for synthesis researchers and other laboratory users to analyze their own samples. It provides an effective tool for customers, such as those described below, that like to use the instrument in a manner generally referred to as “open access.”
- Customers who intend to share a single LCMS-IT-TOF system among many researchers, so each can confirm the molecular weight of synthesized compounds or confirm their purity level or contaminants
- Users that want to utilize formula prediction software to confirm synthesis and improve the accuracy of such determinations
- Users with their desks located away from the instruments, which involves time-consuming movement before they can view the results.
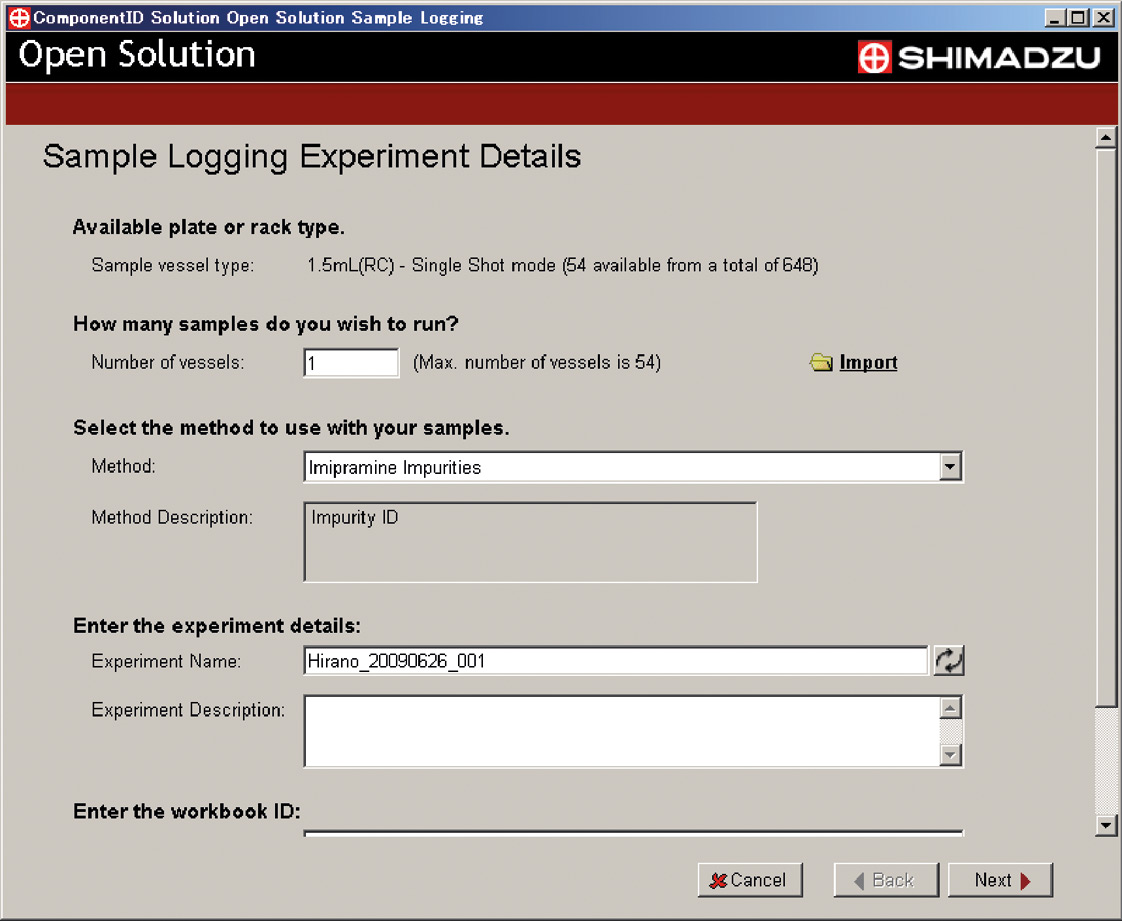 Figure 6b: Login Window of ComponentID Solution
Figure 6b: Login Window of ComponentID Solution
ComponentID offers the following features to address the customers’ needs described above.
- After logging in to the software, it takes only five steps to start an analysis: enter the number of samples, select a method, enter the target chemical composition, place the samples in the autosampler, and click the [Start] button. A report of the results can be received via email. Training is not therefore necessary for general users. LC/MS system administration and method development is performed by the system administrator. The sample logging function is the same as that used in the highly acclaimed Open Solution and PsiPort software for LCMS-2020/LCMS-2010 series systems.
- MSn accurate mass measurement results from the LCMS-IT-TOF system are utilized to accurately predict the composition of synthesized substances and impurities. These results are then output automatically in a report.
- ComponentID supports high-throughput LC/MS analysis using prominence systems, and also supports flow injection analysis which does not use a column. Using flow injection allows the processing of a larger number of samples in a short time.
- Data entry time can be reduced by importing a list of sample information, such as the composition of synthesized compounds, from an Excel spreadsheet or directly copying and pasting from Excel. This is also convenient for synthetic substances in plate format.
- On completion of an analysis, obtained results are printed as a PDF file, attached to an email and sent to a pre-specified e-mail address. The layout of the output can be customized by the system administrator, so the PDF file can include not only formula prediction software results, but also MS chromatograms, spectra, LC chromatograms or other information desired by the user.
- In addition, ComponentID includes many other functions developed from actual operation of OpenSolution and PsiPort systems, such as a desktop lock feature, overnight analysis feature and priority settings.