Hair in your soup?
Microchip electrophoresis

The production of foods, drugs, cosmetics etc. demands high standards of purity in produced goods. It is essential to rule out all types of contamination. If contamination becomes evident despite all precautionary measures taken, it is necessary to discover and identify the source quickly, reliably and accurately. Only in this way can a standard procedure be developed to prevent the occurrence of future incidents.
Contaminations are usually ascertained by microscopic analysis which requires extensive experience and specialized knowledge of the staff, particularly when identifying contaminations by animal hair. A reliable and fast alternative is the identification of animal species by polymerase chain reaction (PCR) with subsequent analysis using microchip electrophoresis.
The PCR technique enables fast replication of double-stranded DNA. Using a specific primer, it is possible to synthesize DNA fragments that are specific to different animal species. Analysis and assignment of these fragments is carried out on the basis of their size.
To determine the size of the DNA fragments, agarose gel electrophoresis following PCR is conventionally used. An electric field is applied to the agarose gel so that the negatively charged DNA migrates through the gel. The agarose gel forms a net-like matrix in which the DNA fragments are separated in proportion to their size. They are then stained using a fluorescent dye (usually ethidium bromide [EtBr]) and visualized under UV light.
Modern automated method
A modern alternative to conventional agarose gel electrophoresis is Shimadzu’s automated microchip electrophoresis system “MCE-202 MultiNA” (Figure 1). Size determination of DNA fragments as well as quantification is achieved using microchip technology. The reusable quartz microchips (Figure 2) have a 23 mm long separation channel in which the DNA fragments are separated in a polymer separation buffer.
 Figure 1: Shimadzu’s fully automated MCE 202 MultiNA microchip electrophoresis system
Figure 1: Shimadzu’s fully automated MCE 202 MultiNA microchip electrophoresis system
 Figure 2: Reusable quartz microchip
Figure 2: Reusable quartz microchip
Filling of the microchips with separation buffer and sample, carrying out of electrophoresis and subsequent rinsing are combined in a fully automated process. A total of four microchips can be installed in the instrument. Using multiple microchips simultaneously, the parallel workflow (rinsing, loading, electrophoresis) enables a reduction in cycle time to 75 seconds per sample.
Up to 120 samples per run can be registered in an analysis plan. In addition to 96-well plates, suitable sample vials are 8-strip or 12-strip tubes or single tubes. Size calibration curves with individually selectable DNA ladders are recorded for each microchip. These calibration curves enable accurate determination of the fragment sizes in the samples.
To compensate for small production-related variation between microchips, the system automatically adds an internal size marker system (lower marker and upper marker) to the samples prior to each analysis. The lower and upper markers restrict the size range that can be analyzed. In addition to the internal marker system, the size range that can be analyzed is also determined by the separation buffer used. Separation buffer and internal markers are sold together as a reagent kit. Currently, by selecting one of four reagent kits for double-stranded DNA, a size range of 25 base pairs up to 12,000 base pairs is covered. The RNA kit covers a size range up to 5,000 nucleotides (Figure 3).
 Figure 3: Four reagent kits for size determination of DNA and one reagent kit for RNA analyses
Figure 3: Four reagent kits for size determination of DNA and one reagent kit for RNA analyses
Advantages of the automated method
Automated microchip electrophoresis offers numerous advantages over conventional gel electrophoresis:
- High sensitivity – the fluorescent stain used and the optical detection system are up to ten times more sensitive than conventional ethidium bromide staining
- Fast fully automated operation – parallel processing by a total of four microchips can reduce the cycle time to 75 seconds per sample
- High reproducibility i. a. due to the low number of manual steps and the associated reduction in sources of error
- Easy operation via straightforward and well designed software
- Low running costs through reusable microchips
When combining a PCR-based verification with subsequent detection using the MultiNA, an automated, fast and reliable method for the detection of contamination by animal hair can be obtained. Hair of six different farm animals (cow, pig, chicken, horse, sheep and goat), three pets (dog, cat and rabbit) as well as three rodents (sewer rat, black rat and house mouse), has been unequivocally identified (Figure 4). DNA of the corresponding animals was extracted from their hair and used for PCR analysis. Specific primer sets generating PCR fragments of specific sizes for each animal were used.
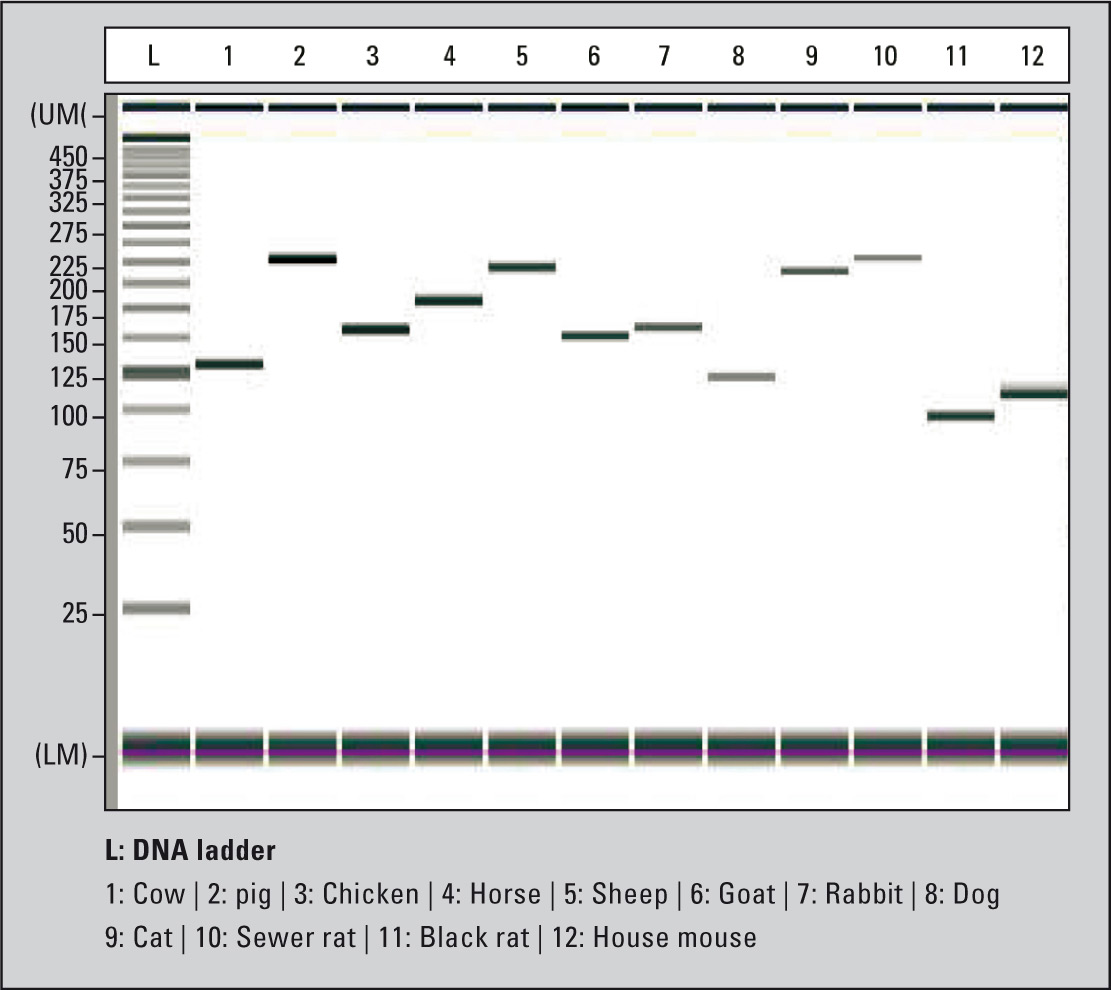 Figure 4: Identification of specific PCR products of different animal species using the MultiNA
Figure 4: Identification of specific PCR products of different animal species using the MultiNA
Following the PCR, size determination of PCR fragments was carried out using the MultiNA. In this way, it is possible not only to discover contaminations that have occurred, but also to unequivocally assign them and to identify their source (for instance farm or domestic animals). Only this will ensure that the source of the contamination can be reliably excluded and that, in future, no one finds a hair in the soup, pain reliever or skin cream.