Holstein and Slovak spotted cattle breeds
Detection of DGAT1 gene polymorphism using microchip electrophoresis
By M. Bauer1,2, D. Vašiček1 and K. Vašičkova1
1 Animal Production Research Centre, Nitra, Slovakia
2 Constantine the Philosopher University, Nitra, Slovakia

The aim of this study was to detect K232A polymorphism in acyl-CoA:diacylglycerol-acyltranferase-1 gene (DGAT1) in the Holstein and Slovak spotted cattle breeds in Slovakia using improved primers and high-throughput microchip electrophoresis (MCE).
Samples of 87 animals of Holstein breed and 42 animals of Slovak spotted breed were genotyped for DGAT1 K232A polymorphism (A and K alleles) using the PCR-RFLP technique.
Clear resolution of three different genotypes was observed following fast, automated analysis of PCR products by microchip electrophoresis. The Holstein breed showed frequencies of 0.8621 for the A allele and 0.1379 for the K allele.
Most animals were AA homozygotes (frequency 0.7356) or AK heterozygotes (frequency 0.2529). Only one KK homozygote (0.0115) was found in the tested sample. The observed frequency of the allele K in tested herds of Slovak Holstein is lower than reported for this breed in other countries. Genotyping data for K232A polymorphism in the Slovak spotted breed showed even lower frequency of the allele K (0.0476) than in the Holstein breed. The results could be used to guide further association studies between this locus and milk traits in these breeds.
Introduction
DGAT1 encodes the enzyme acyl-CoA:diacylglycerol-acyltransferase that plays a fundamental role in the metabolism of cellular diacylglycerol in physiological processes such as intestinal fat absorption, lipoprotein assembly, adipose tissue formation and lactation involved in the metabolism of triacylglycerol in higher eukaryotes [1]. Additional studies have demonstrated the effect of K232A mutation (substitution of lysine by alanine at position 232 of DGAT1 protein) on the function of the resulting enzyme [2]. Other studies have shown that the 2 alleles at the K232A polymorphism segregate in several breeds from different countries [3].
Nevertheless, the estimated frequency of alleles varies greatly according to the population considered, which may reflect different breeding objectives regarding milk composition in different countries and breeds. A significant decrease in protein and milk yields, and increase in fat yield was associated with the lysine substitution (K allele). The alanine variant (A allele) was associated with an increase in protein and milk yields and decrease in fat yield [4;5].
Microchip electrophoresis has recently attracted much attention in DNA analysis due to its high efficiency, high throughput, time-saving ability, easy operation and low consumption of samples and reagents [6]. In microchip electrophoresis (MCE), nucleic acid fragments are separated at high voltage by capillary electrophoresis in a chip with micro-fabricated channels, with automated detection as well as on-line data evaluation. MCE has therefore emerged as an effective tool for the electrophoretic separation of a variety of different DNA inputs while providing a viable alternative to conventional agarose or PAGE slab gel electrophoretic applications.
Materials and methods
DNA from hair roots and blood was isolated by the Maxwell 16 Magnetic Particle Processor and corresponding tissue or blood Maxwell purification kit (Promega, USA) following the manufacturer’s instructions. Genomic DNA was genotyped by an PCR-RFLP assay for the locus responsible for the DGAT1 K232A substitution. Briefly, PCR reactions were performed in a total volume of 20 µL using 10-50 ng of genomic DNA as template, 1X PCR buffer, 2.5 mM MgCl2, 0.2 mM of each dNTP, 5 % DMSO, 0.8 U AmpliTaq Gold DNA polymerase (Applied Biosystems, USA) and 0.4 µM of each primer. The PCR profile included an initial denaturation step at 95 °C for 5 min, 35 cycles of 94 °C (60 s), 63 °C (60 s) and a final extension step of 10 min at 72 °C. Modified primers for the amplification of 352 bp fragment of bovine DGAT1 gene (sequence acc. No. AY065621.1; dgat1 forward: 5’-catcctcttcctcaagctgttct-3’; dgat1 reverse: 5’-gggcgaagaggaagtagtagaga-3’) were designed using Primer3 software. Restriction endonuclease CfrI (Fermentas, Germany) was used to digest a 352 bp PCR product. The uncut fragment represents the lysine variant, whereas the CfrI fragments of 199 and 153 bp represent the alanine variant.
PCR products were analyzed using the automated MCE®-2020 MultiNA microchip electrophoresis system (Shimadzu, Japan) with a DNA-500 kit according the manufacturer’s protocol. An SYBR Gold fluorescent dye for DNA staining (Invitrogen, USA) and a 25 bp DNA Ladder (Invitrogen, USA) was used to determine the size of the PCR products.
A software PowerMarker v.3.25 [7] was used to calculate allelic and genotypic frequencies and to estimate the deviation from Hardy-Weinberg equilibrium for a given locus.
Results and discussion
87 animals of Holstein breed and 42 animals of Slovak spotted breed have been tested for K232A polymorphism in DGAT1 gene using novel primers for PCR-RFLP genotyping. New primers have been designed generating the 352 bp PCR product. The uncut fragment represents the lysine variant (K allele), whereas the CfrI fragments of 199 bp and 153 bp represent the alanine variant (A allele). The digestion products could be analyzed effectively by fast, automated and high-throughput microchip electrophoresis showing a clear separation of three different genotypes (Figure 1).
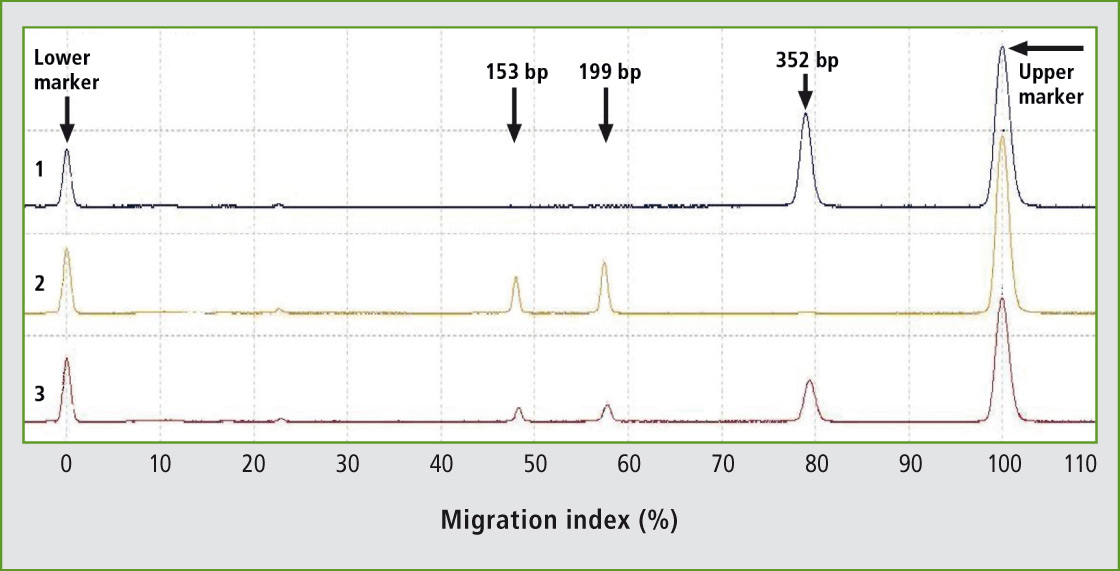 Figure 1: Representative results of PCR-RFLP genotyping of K232A polymorphism of DGAT1 gene in the Holstein breed using the microchip electrophoresis (MCE). Lane 1: genotype KK; Lane 2: genotype AA; Lane 3: genotype KA.
Figure 1: Representative results of PCR-RFLP genotyping of K232A polymorphism of DGAT1 gene in the Holstein breed using the microchip electrophoresis (MCE). Lane 1: genotype KK; Lane 2: genotype AA; Lane 3: genotype KA.
The allele and genotype frequencies as well as the estimation of Hardy-Weinberg equilibrium were calculated by PowerMarker v.3.25 and are summarized in Table 1. The observed frequency of the allele K in the Slovak Holstein cattle herd tested is lower than reported for this breed in the other countries. In German Holstein allele frequencies reported for the lysine variant K range from 0.35 to 0.548 depending on the sample [4], while French Holstein showed a frequency of K allele of 0.37 [9].
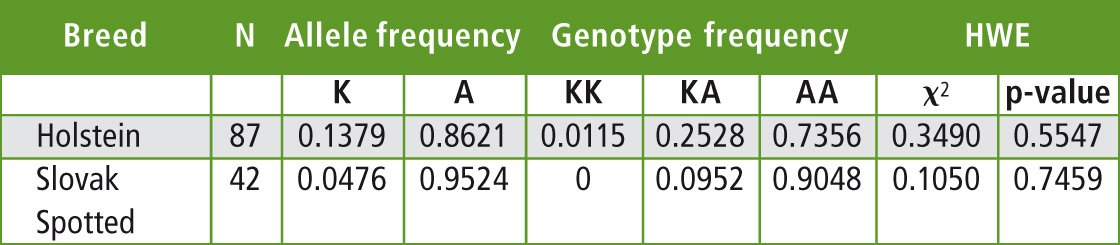 Table 1: The allele and genotype frequencies and Hardy-Weinberg equilibrium of DGAT1 K232A polymorphism in the tested population of Holstein and Slovak spotted breeds
Table 1: The allele and genotype frequencies and Hardy-Weinberg equilibrium of DGAT1 K232A polymorphism in the tested population of Holstein and Slovak spotted breeds
The data regarding K232A polymorphism of DGAT1 gene in the Slovak spotted breed showed even lower frequency of the allele K (0.0476) than in the Holstein breed. 38 AA homozygotes (frequency 0.9048) were found, but only four AK heterozygotes (0.0952) in 42 tested animals. Such a low frequency was also reported in the spotted cattle breeds [4; 8]. The chi-square test found a deviation from the Hardy-Weinberg equilibrium of the DGAT1 K232A genotypic frequencies for Holstein and Slovak spotted breeds. This disequilibrium could be related to the sampling of the genotyped animals or can be a result of indirect selection for this locus from the selection for milk production.
The results could be used to guide further association studies between this locus and milk traits, e.g. milk production, fat and protein content in the milk. Well-established diagnostics of DGAT1 variants in individual animals might be extremely helpful to further investigate biochemical pathways involved in the expression of milk production traits or to study possible interactions with other causal genes.
Acknowledgements
This article was written during realization of the “LAGEZ No. 26220120051” and “MARKERY No. 26220220033” projects supported by the Operational Programme Research and Development, funded through the European Regional Development Fund.
References
- Cases S., Smith S.J., Zheng Y.W., Meyers H.M., Lear S.R., Sande E., Novak S., Collins C., Welch C.B., Lusis A.J., Erickson S.K., Farese R.V.; 1998. Identification of a gene encoding an acylCoA: diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA, vol. 95, 1998, p. 13018-13023
- Grisart B., Farnir F., Karim F., Cambisano N., Kim J.J., Kvasz A., Mni M., Simon P., Frere J.M., Coppieters W., Georges M.; 2004 Genetic and functional confirmation of the causality of the DGAT1 K232A quantitative trait nucleotide in affecting milk yield and composition. Proc. Natl. Acad. Sci. USA, vol.101, 2004, p. 2398-2403
- Kaupe B., Winter A., Fries R., Erhardt G.; 2004. DGAT1 polymorphism in Bos indicus and Bos taurus cattle breeds. J. Dairy Res., vol. 71, no. 2, 2004, p. 182-187
- Thaller G., Krämer W., Winter A., Kaupe B., Erhardt G., Fries R.; 2003. Effects of DGAT1 variants on milk production traits in German cattle breeds. J. Anim. Sci., vol. 81, 2003, p. 1911-1918
- Weller J.I., Golik M., Seroussi E., Ezra, E., Ron M.; 2003. Populationwide analysis of a QTL affecting milkfat production in the Israeli Holstein population. J. Dairy Sci., vol. 86, 2003, p. 2219-2227
- Zhang L., Dang L., Baba Y.; 2003. Microchip electrophoresis – based separation of DNA. J. Pharm. Biomed. Anal., vol. 30, 2003, p. 1645-1654
- Liu K., Muse S.V.; 2005. Power Marker: An integrated analysis environment for genetic marker analysis. Bioinformatics, vol. 21, 2005, p. 2128-2129
- Winter A., Kramer W., Werner F.A., Kollers S., Kata S., Durstewitz G., Buitkamp J., Womack J.E., Thaller G., Fries R.; 2002. Association of a lysine-232/alanine polymorphism in a bovine gene encoding acyl-CoA:diacylglycerol acyltrans-ferase (DGAT1) with variation at a quantitative trait locus for milk fat content. Proc. Natl. Acad. Sci. U.S.A., vol. 99, 2002, p. 9300-9305
- Gautier M., Capitan A., Fritz S., Eggen A., Boichard D., Druet T.; 2007. Characterization of the DGAT1 K232A and variable number of tandem repeat polymorphisms in French dairy cattle. J. Dairy Sci., vol. 90, 2007, p. 2980-2988